Onboarding
Software to install
- Visual Studio Code: Code editor for HTML/JS/CSS and Markdown. Download
- R: Statistical computing for data cleaning and analysis. Download
- R Studio: Provides a basic interface to R. Download
- Microsoft Teams: Lab communication and meetings. Download
- OneDrive (desktop app): Local sync of your lab folders with version history. Download
- EndNote: Reference manager with shared lab library. Download
- GitHub Desktop: Simple Git client for version control. Download
- JASP: Point-and-click stats (Bayesian and frequentist). Download
- Zoom: Online meetings and participant calls. Download
Accounts to set up
- A MetaForge account. You can join the MetaLab team using code MTVJ6K
- A SONA account (if using SONA). Apply to be added as a new user here.
- A Prolific account (if using Prolific).
- A Github account using the USYD enterprise version. To sign up you simply need to login. To access some of secure lab resources you will need to be added to the USYD Meta Lab Github Enterprise team (contact Kit with your unikey).
- A LabArchives Scheduler account to book the lab testing space. See the School intranet.
- You need to have been added to one of the lab ethics protocols. Honours students will need to manually apply to be added as a user on myResearch Ethics before being added.
Communication
Face-to-face communication is preferred whenever possible, either during our weekly one-on-one meetings or in the group lab meeting. For quick questions, please feel free to drop by if my door is open. Email should be reserved for more formal communication or for tasks that require detailed input, for example, requesting feedback on a draft manuscript or thesis section.
Larger multi-lab meetings focused on either cognition (SLAC) or emotion (REAL) also occur regularly and students are encouraged to attend.
Folders
Project folder structure
Projects follow a standard structure to keep code, analyses, and outputs tidy.
Project_Name/
├── README.md # Project overview and folder structure
├── .gitignore # Files to ignore in version control
├── experiments/
│ └── example-experiment/
│ ├── design/ # Task design files, protocols
│ ├── stimuli/ # Images, sounds, or other stimuli
| ├── preregistration/ # Preregistration documents
│ ├── scripts/ # Code for running the experiment (e.g., jsPsych, MATLAB)
│ ├── data/
│ │ ├── metadata/ # Data dictionary and metadata files
│ │ ├── processed/ # Cleaned data ready for analysis
│ │ └── raw/ # Untouched raw data
│ ├── analysis/
│ │ ├── scripts/ # R, Python, MATLAB analysis scripts
│ │ ├── results/ # Figures, stats outputs, tables
│ │ ├── notebooks/ # RMarkdown or Jupyter notebooks
│ │ └── analysis_plan.Rmd # Pre-planned analysis description
│ ├── code/ # Shared functions or custom packages for this experiment
│ └── notes.md # Lab notes or changelog for the experiment
├── manuscripts/
├── reports/ # Internal reports or summaries
└── presentations/ # Conference slides, posters, etc.
Tip: Use the lab R package function to produce a project structure quickly:
MetaLab::create_MetaLab_project("MyProject", "MyExperiment", "Directory")
Launching a study
Running a Study
Step-by-Step Guide to Launching a Study
🔹 Step 1: Build Your Experiment
- Build your experiment on MetaForge
- For help with building your experiment, use the built ion Gemini AI or if you have a paid Claude account use the bot
- Click the Publish button (in the top toolbar) to copy the experiment link to your clipboard
- The URL format is:
https://metaforge.live/?run={your-project-id}
- The URL format is:
🔹 Step 2: Putting the study on Prolific
Option A: Use MetaForge's Prolific Integration
- Open Project Settings (gear icon) → Prolific tab
- Enter your Prolific API Key (from Prolific > Settings > API Tokens)
- Fill in the study details (name, description, duration, reward, etc.)
- Click Create Draft Study — this will create a draft directly on Prolific
Option B: Manual Setup on Prolific
- Go to Prolific
- To be added to the lab team (and use lab funds), let Kit know your Prolific username
- Create a new study:
- Study name and Study description: from ethics documents
- Internal study name: use format
LastName-TASK-experiment-name - Device compatibility: typically limit to desktop only
- Study URL: use the URL from Step 1. Append the Prolific ID using URL parameters:
- Select Recording Prolific IDs = URL parameters
- Final URL should look like:
https://metaforge.live/?run=YOUR_PROJECT_ID&PROLIFIC_PID={{%PROLIFIC_PID%}} - Use
&(not?) if you already have parameters in the URL
- Set completion paths and copy the completion code (e.g.,
CGJ91R8S) - In MetaForge, go to Project Settings → Redirects and set the Default Redirect URL to:
https://app.prolific.com/submissions/complete?cc=YOUR_COMPLETION_CODE - Set duration and pay participants £6/hour
- Set sample country and use Quota Sample for balanced gender distribution
🔹 Step 3: Putting the study on SONA
- Read the SONA handbook for detailed procedures
- Go to SONA
- Select Add new Study → Online External Study
- Fill in details from ethics documents
- Study name, Brief Abstract, Detailed Description
- Duration and credits (1 credit = 60 minutes)
- Human Res Ethics Com Approval Code (e.g.,
2022/796)
- Study URL: append the SONA ID:
https://metaforge.live/?run=YOUR_PROJECT_ID&SONAID=%SURVEY_CODE% - Set Active Study? = Yes and Automatic Credit Granting = Yes
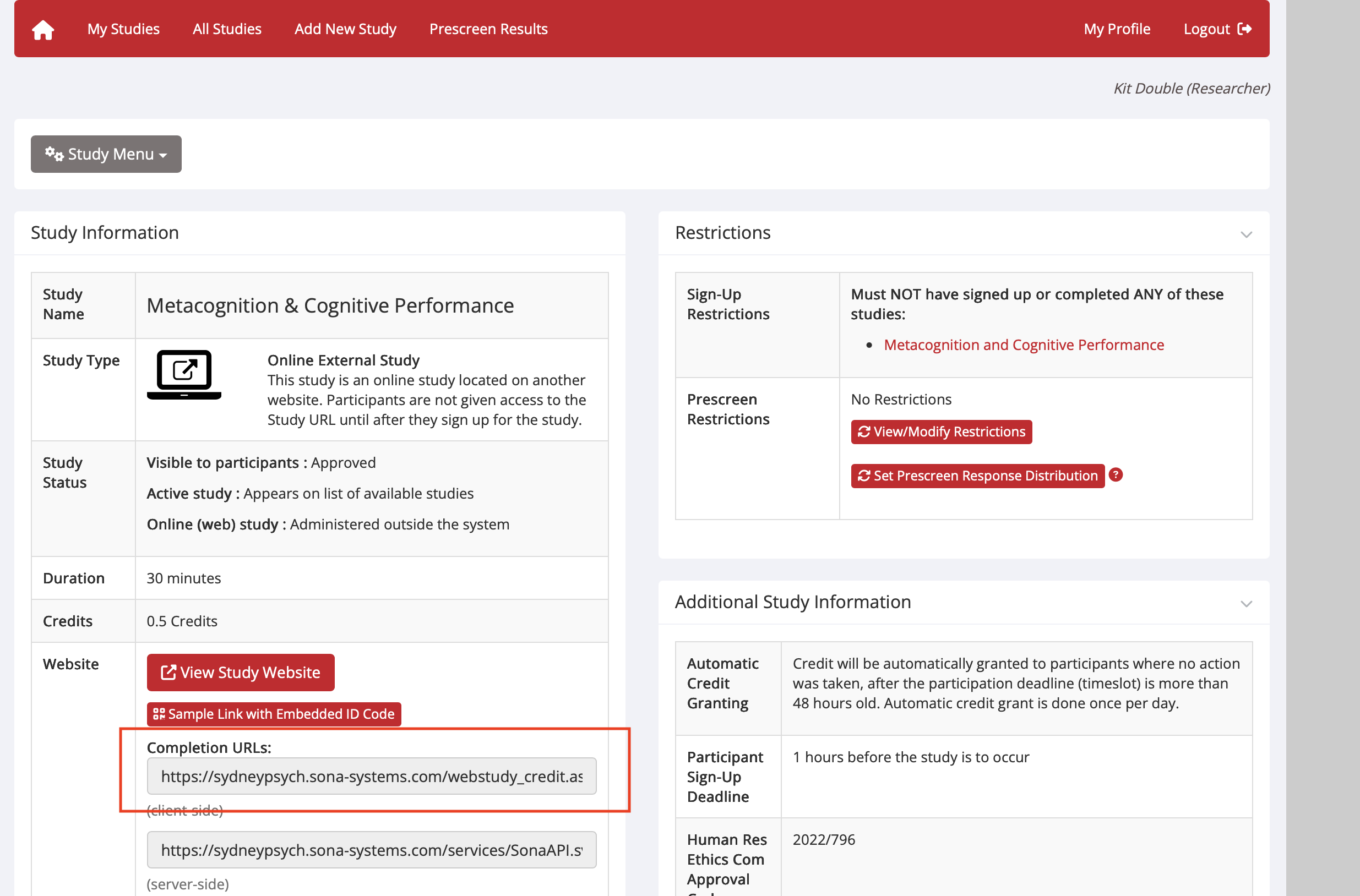
- Copy your SONA experiment ID and credit token into the redirect URL. See highlighted section below:
- More info on SONA redirect
- In MetaForge, go to Project Settings → Redirects and set the Default Redirect URL to your SONA completion URL. To find this:
- Go to SONA → Study Information → Change Study Information
- Find the completion URL format (includes your experiment ID and credit token)
- Request approval for your SONA study from School research staff
- For the ethics description question, copy from the ethics documents
🔹 Step 4: Test the Study
- Visit your experiment link
- Add test parameters to simulate real participants:
- Prolific:
https://metaforge.live/?run=YOUR_PROJECT_ID&PROLIFIC_PID=123ABC - SONA:
https://metaforge.live/?run=YOUR_PROJECT_ID&SONAID=S1234567
- Prolific:
- Run through the study yourself
- Confirm:
- Data saves to MetaForge (check participant count)
- Redirect works correctly at the end
🔹 Step 5: Preregistration
- If preregistering, do so before collecting data
- Recommended platform: AsPredicted
🔹 Step 6: Go Live!
- Launch on SONA (add slots) or Prolific (publish study), or both
- Monitor incoming data on your MetaForge project dashboard
- Download data directly from MetaForge, or use the R API:
# First, source the MetaForge R functions by loading the lab R package library(MetaLab) # Set your API token (get this from your Profile in MetaForge) metaforge_set_token("your-api-token-here") # List all your projects projects <- metaforge_list_projects() print(projects) # Download data from a specific project data <- metaforge_get_data("your-project-id") # Or download all project data at once all_data <- metaforge_get_all_data(combine = TRUE)